Azolla microphylla (Mexican Mosquito Fern)
Visual overview of genome assembly metrics
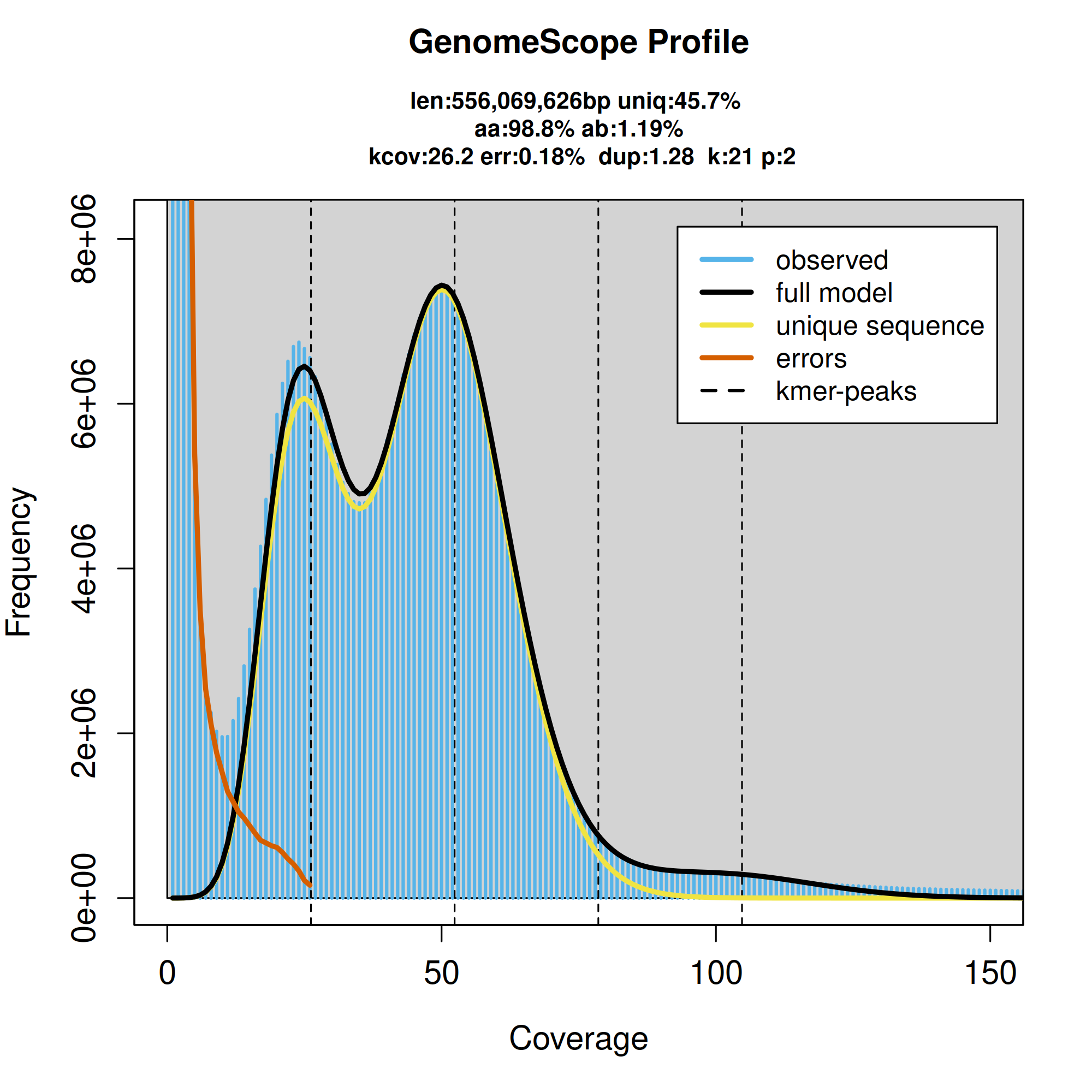
K-mer spectra output generated from PacBio HiFi data without adapters using GenomeScope2.0. The bimodal pattern observed corresponds to a diploid genome and the k-mer profile matches that of high heterozygosity.
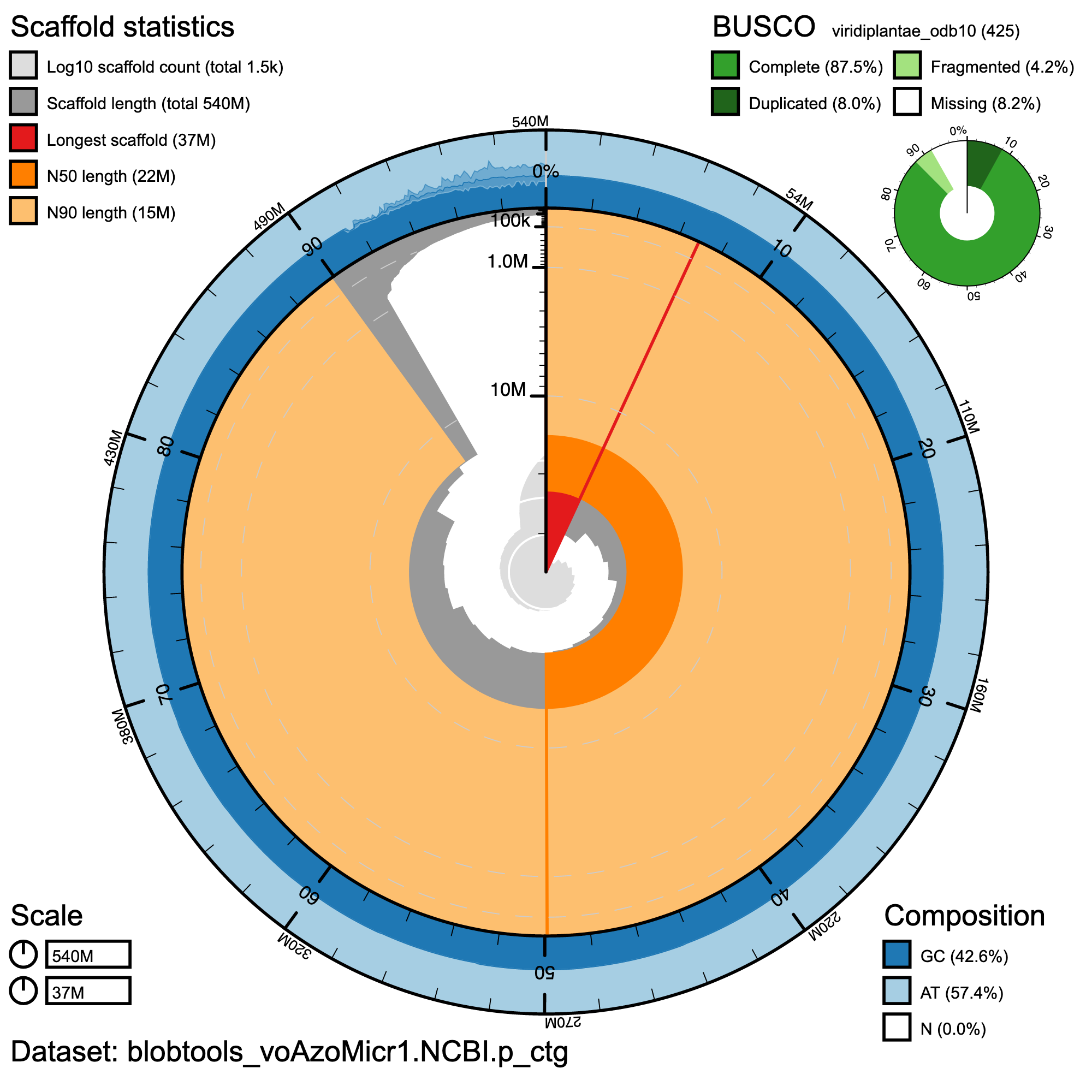
BlobToolKit snail plot showing a graphical representation of the quality metrics for haplotype 1. The first diagonal (~30 °C) line represents the size of the longest scaffold; all other scaffolds are arranged in order of size clockwise around the plot and drawn in gray starting from the outside of the central plot. The inner arcs show the scaffold N50 and scaffold N90 values. The central light gray spiral shows the cumulative scaffold count with a white line at each order of magnitude. White regions in this area reflect the proportion of Ns in the assembly; the outer dark vs. light area around it shows mean, maximum, and minimum GC vs. AT content at 0.1% intervals (Challis et al. 2020).
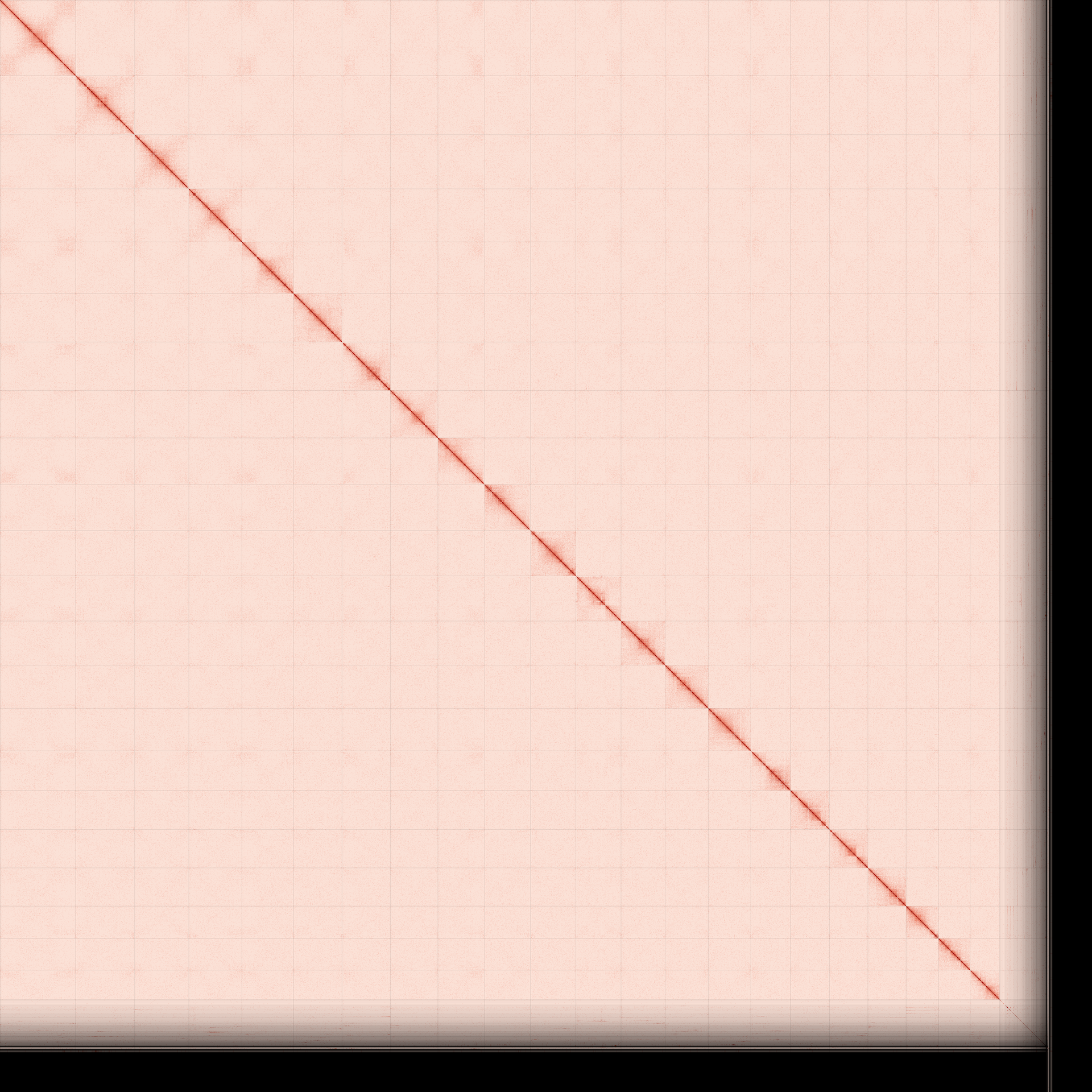
Omni-C contact maps for the curated genome assembly of haplotype 1 generated with PretextSnapshot. Omni-C contact maps translate the proximity of genomic regions in 3D space to contiguous linear organization. Each cell in the contact map corresponds to sequencing data supporting the linkage (or join) between 2 of such regions. Scaffolds are separated by black lines.
Authors
Michael J Song, Yanã C Rizzieri, Fay-Wei Li, Forrest Freund, Merly Escalona, Erin Toffelmier, Courtney Miller, H Bradley Shaffer, Oanh Nguyen, Mohan P A Marimuthu, Noravit Chumchim, Carrie Tribble, Colin W Fairbairn, William Seligmann, Carl J Rothfels