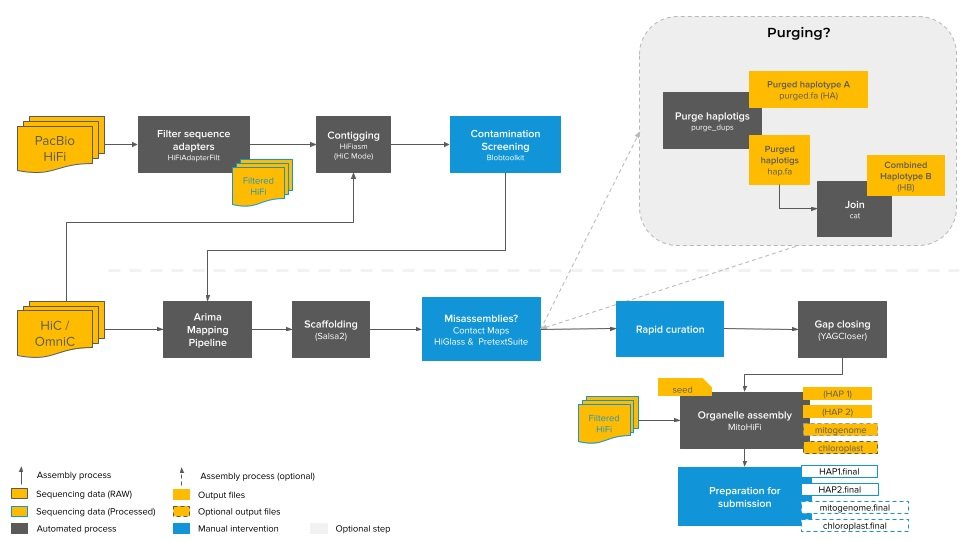
CCGP reference genomes are assembled following a protocol adapted from Rhie et al. (2021). Assemblies are comprised of PacBio HiFi long read data, which is scaffolded using Omni-C (Dovetail Genomics) chromatin conformation data. Our minimum target reference genome quality is 6.7.Q40, and in most cases we expect to reach 7.C.Q50 or better (see Table 1 in Rhie et al. 2021).
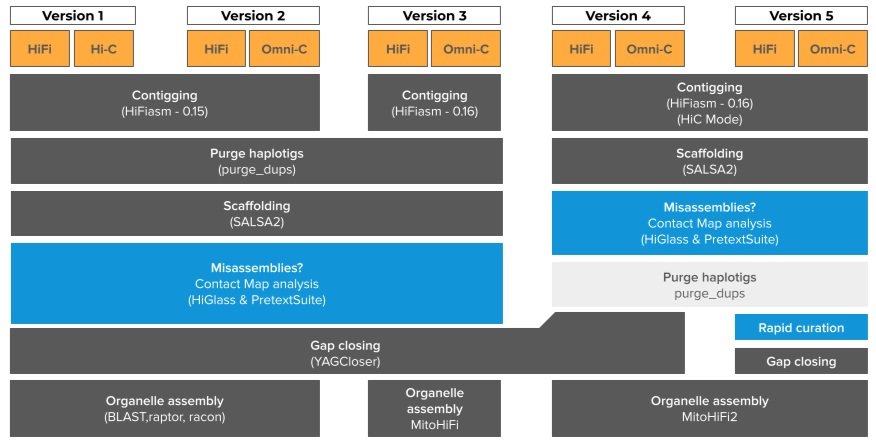
Evolution of the CCGP assembly pipeline
Standard CCGP assembly pipeline
This pipeline is a general schematic and may vary slightly from assembly to assembly
Genome Assembly Software
| Software | Version | |
|---|---|---|
| Assembly | ||
| Filtering PacBio HiFi adapters | HiFiAdapterFilt | Commit 64d1c7b |
| Kmer counting | Meryl | 1 |
| Estimation of genome size and heterozygosity | GenomeScope | 2 |
| De novo assembly (contigging) | HiFiasm | 0.13-r308 |
| Long read, genome-genome alignment | Minimap2 | 2.16 |
| Remove low-coverage, duplicated contigs | Purge_dups | 1.0.1 |
| Scaffolding | ||
| OmniC mapping for SALSA | Arima Genomics mapping pipeline | Commit 2e74ea4 |
| OmniC Scaffolding | SALSA | 2 |
| Gap closing | YAGCloser | Commit 20e2769 |
| Hi-C contact map generation | ||
| Short-read alignment | Bwa | 0.7.17-r1188 |
| SAM/BAM processing | Samtools | 1.11 |
| SAM/BAM filtering | pairtools | 0.3.0 |
| Pairs indexing | pairix | 0.3.7 |
| Matrix generation | Cooler | 0.8.10 |
| Matrix balancing | hicExplorer | 3.6 |
| Contact map visualization | HiGlass | 2.1.11 |
| PretextMap | 0.1.4 | |
| PretextView | 0.1.5 | |
| PretextSnapshot | 0.0.3 | |
| Organelle assembly | ||
| Sequence similarity search | BLAST+ | 2.1 |
| Long read alignment | Pbmm2 | 1.4.0 |
| Variant calling and consensus | bcftools | 1.11-5-g9c15769 |
| Extraction of sequences | seqtk | 1.3-r115-dirty |
| Circular-aware long-read alignment | racon | 1.4.19 |
| Sequence polishing | raptor | 0.20.3-171e0f1 |
| Sequence alignment | lastz | 1.04.08 |
| Gene annotation | MitoFinder | 1.4 |
| Organelle annotation | GeSeq | |
| Genome quality assessment | ||
| Basic assembly metrics | QUAST | 5.0.2 |
| Assembly completeness | BUSCO | 5.0.0 |
| Merqury | 1 | |
| Contamination screening | ||
| General contamination screening | BlobToolKit | 2.3.3 |
The software and software versions listed above may vary slightly from assembly to assembly